New method opens crystal-clear views of biomolecules

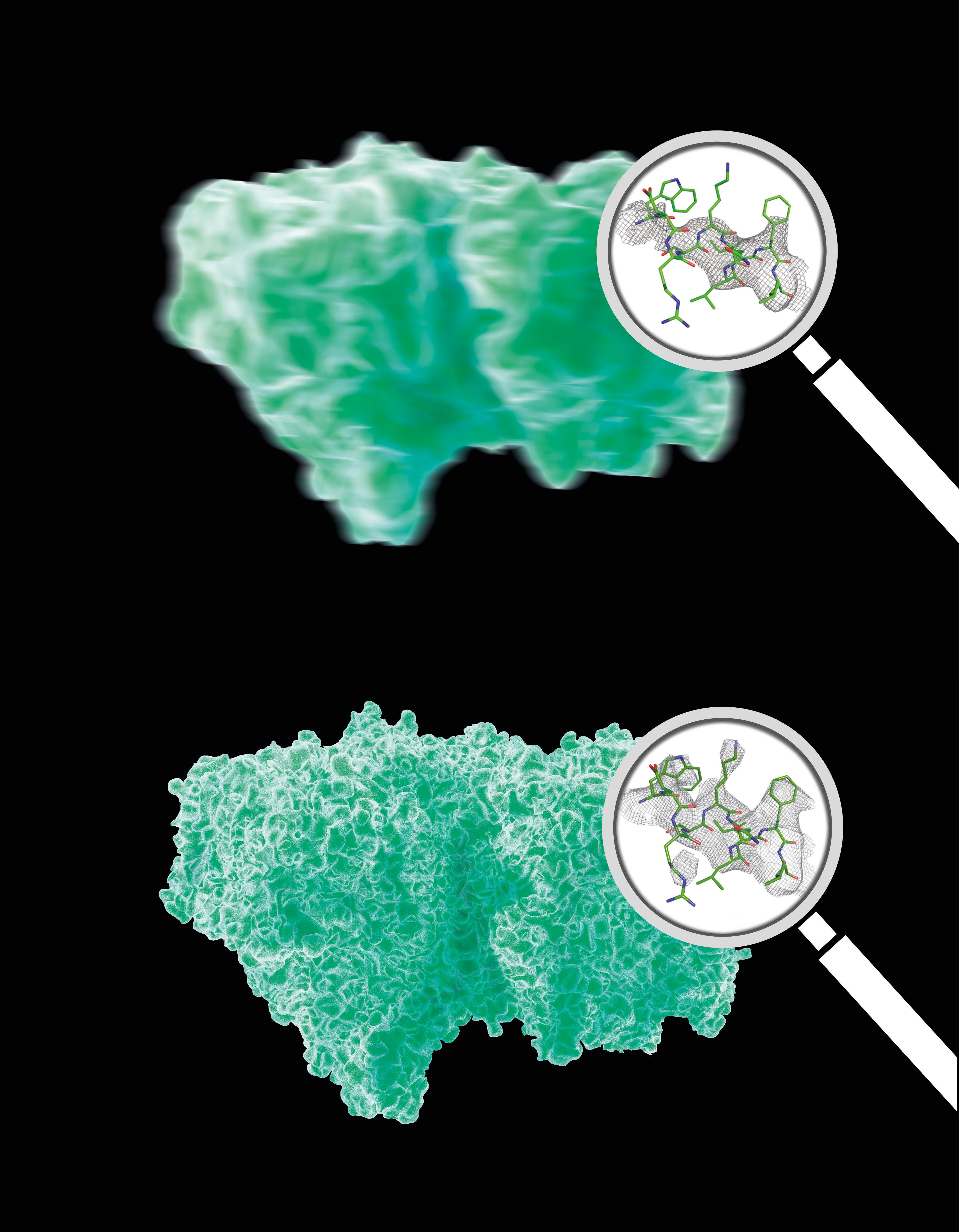
Slightly disordered crystals of complex biomolecules like that of the photosystem II molecule shown here produce a complex continous diffraction pattern (right) under X-ray light that contains far more information than the so-called Bragg peaks of a strongly ordered crystal alone (left). Note that the disorder is much smaller than what is depicted here. The displacement of the PSII dimers (each consist of more than 100,000 atoms) is very small, corresponding to less than the bond length between two carbon atoms. Credit: Eberhard Reiman/DESY
A scientific breakthrough gives researchers access to the blueprint of thousands of molecules of great relevance to medicine, energy and biology. In a new study, researchers from Arizona State University (ASU), Deutsches Elektronen-Synchrotron (DESY) and Stanford Linear Accelerator Laboratory (SLAC) describe a simple way to determine the three-dimensional structures of proteins and other molecules, many of which are inaccessible by existing methods.
The research findings appear in the current issue of the journal Nature.
Imaging the molecular building blocks of living things at the atomic scale is tricky. Often, the most difficult step is getting such molecules to form high-quality crystals needed for X-ray imaging. This international team describes a new method that can produce sharp images relying on crystals with very small imperfections, using the world’s brightest X-ray source at the Department of Energy’s SLAC National Accelerator Laboratory.
“Once the full potential of the new method is understood, it could turn out to be one of the biggest advances since the birth of crystallography,” said Mike Dunne, director of the Linac Coherent Light Source (LCLS) X-ray laser, a DOE Office of Science User Facility.
The structures of biomolecules reveal their modes of action and provide insights into the workings of the machinery of life. Unlocking the molecular structure of particular proteins, for example, can provide the basis for developing tailor-made drugs against numerous diseases or advancing clean-energy technologies with the efficiency of nature and the stability of engineered systems.
The chosen target of the new study is a critical enzyme known as photosystem II. Petra Fromme, co-author of the new study and director of the Biodesign Institute’s Center for Applied Structural Discovery (CASD) at ASU, highlights the importance of this molecule:
“Life on Earth relies directly on photosynthesis — the natural process of converting light energy to chemical energy in plants and algae. The critical first step is carried out by the photosystem II protein complex, which uses sunlight to split abundant sources of water (H2O) into hydrogen, electrons and all the oxygen that sustains our planet,” she said.
In 2011, the team of researchers at ASU (including Fromme and John Spence, professor of physics, along with their collaborators), pioneered a technique called serial femtosecond crystallography. Here, structure determination is based on imaging of thousands of nanocrystals, so small that they can not even be seen under a microscope. The nanocrystals are hit “on-the-fly” by X-ray pulses that are so strong that they destroy any solid material but so short that a diffraction image is obtained before the destruction occurs.
The method was further developed in 2014 and was used to produce the first snapshot images of the water-splitting process in photosysnthesis. It wasn’t clear, however, if the researchers would ever be able to see the dynamics of the process at atomic scale. Such fine detail coupled with dynamic imaging is essential for understanding the mechanisms of photosynthesis that split water and produce oxygen with visible light and earth abundant metals. Traditional X-ray crystallography methods could achive high resolution but suffer from damage from the X-rays and are not suitable to see dynamics of the process.
“This new method allows us to significantly improve the resolution of the images we took of photosystem II and gives us a new tool for capturing the dyanmics of some of life’s most important processes. When we understand how photosynthesis works at the chemical level we can use this as a blueprint for developing clean and renewable energy technologies,” Fromme added.
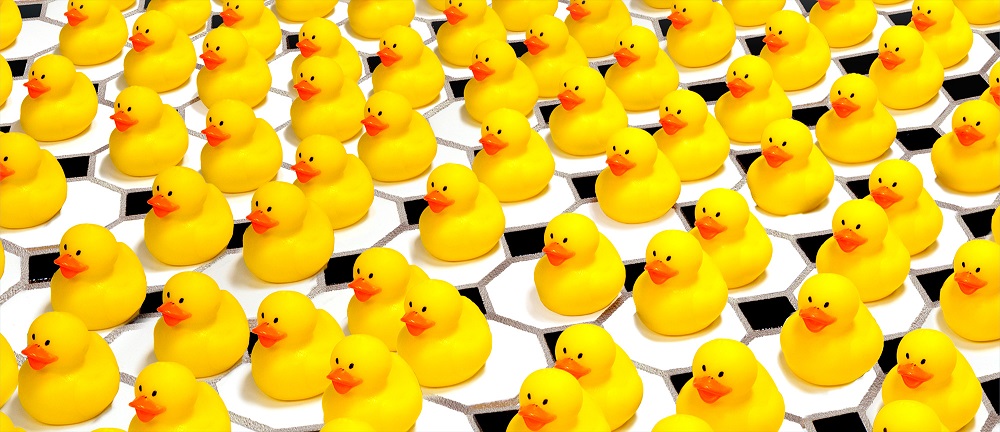
Figure A - Getting the ducks in a row: Artistic representation of an imperfect crystal, in which individual repeating units (represented here by ducks rather than biomolecules) are displaced from their ideal positions on a regular crystal lattice (tiles). In X-ray crystallography, researchers shine X-rays through crystals to determine the atomic structure of the crystals’ units, such as complex biological molecules. A new study of imperfect crystals at SLAC’s LCLS X-ray laser has shown that the imperfections can be exploited to obtain much higher-resolution images than with conventional methods. Credit: SLAC National Accelerator Laboratory
'The best crystals are crystals with small imperfections'
More than 100 years ago, Australian-born British physicist William Lawrence Bragg found a way to use X-rays to probe the interior of crystals, which consist of regular arrays of atoms or molecules. This discovery launched the field of X-ray crystallography, one of the most important techniques for analyzing the structures of materials, chemical processes and biological molecules.
When X-rays pass through the crystal, they scatter off the protein molecules and form a pattern on a detector. This diffraction pattern is dominated by bright spots known as Bragg peaks, which researchers use to reconstruct the atomic structure of the molecules.
Figure B - When X-rays (yellow beam) pass through a crystal (green shapes), they form an intensity pattern on a detector behind the crystal that is dominated by bright spots (dots in the background). Researchers use these so-called Bragg peaks to reconstruct atomic-resolution images of the molecules inside the crystals. In a new study at SLAC’s LCLS X-ray laser, researchers used continuous diffraction — signals found between the spots, which appear here as washed-out, continuous lines in the background — to improve the resolution of images obtained with the conventional analysis. Credit: SLAC National Accelerator Laboratory
A perfectly ordered crystal would produce nothing but Bragg peaks. However, disorder limits the number of detectable peaks and therefore the resolution of the molecular image that can be obtained from the peaks alone.
The small displacements lead to “termination of the Bragg peaks” but allow scientists to “see” the scattering from the sum of the individual molecules in the crystals. These tiny molecular displacements from a perfectly ordered crystal arrangement produce gently rippling patterns between and beyond the sharp Bragg peaks. Although these patterns, known as “continuous diffraction,” have been actively studied, they had not been considered capable of producing high-resolution molecular images.
“We’ve now demonstrated that we can actually use the continuous diffraction of imperfect crystals to obtain better molecular images than with Bragg peaks alone,” said Kartik Ayyer, the study’s first author from the Center for Free-Electron Laser Science (CFEL) at the German research center DESY.
'Phased in'
Even when good diffraction is available, the structure of a complex molecule is still extremely difficulty to determine. Without knowing the phase — the lag of the crests of one diffracted wave to another — it is not possible to compute an image of the molecule from the measured diffraction pattern.
To solve the tricky phase puzzle, more information must be known than just the intensity of the measured Bragg spots. Sometimes this information can be derived by X-analysis of crystals of chemically modified molecules, or inferred from the structure of a closely related molecule, but these approaches are time-consuming and can bias the results. Bragg peaks are strong, since they arise from constructive interference (like a crowd clapping in unison), but they are sparse and of limited content. The continuous diffraction used in the new method fills in the gaps in and beyond the Bragg peaks, giving vastly more information, which can be used to directly obtain the phase and improve the resolution.
ASU physicist and study co-author John Spence elaborated:
“In 1913, Paul Debye showed that Bragg diffraction from vibrating atoms in a crystal scatter between the Bragg spots. Henry Chapman (XFEL scientist with DESY and corresponding author) was able to show how, in a similar way, the non-Bragg scattering from LCLS X-ray snapshots from imperfect crystals can be used to provide a resolution-enhanced image of the protein molecule, and also solve the phase problem."
This simple concept leads to a paradigm shift in crystallography — the most ordered crystals are no longer the best to analyze. Imperfect crystals arise because some molecules do not align perfectly with majority of the molecules within the crystal (see Figure A), which creates a continuous diffraction pattern similar to those predicted for single molecules. Single molecule diffraction is the Holy Grail of modern X-ray science because it eliminates the crystallization process altogether, but has never been possible because the signals from individual molecules are far too weak.
The analysis of the Bragg peaks alone (top) reveals far less detail than the analysis of the continuous diffraction pattern (bottom). Magnifying glassses show real data. Credit: Eberhard Reimann/DESY
“For the first time we have access to single molecule diffraction — we have never had this in crystallography before,” Chapman said. The technique elegantly weds X-ray diffraction of crystals and X-ray imaging of single particles, providing the best of both worlds.
The technique could provide a stepping-stone toward detailed imaging of single particles, including the large number of biological specimens that cannot easily be crystallized
Fromme stated that the new technique also opens the field for time-resolved movies of biomolecules at work. “While perfect crystalline order restricts the movement of molecules, the new method allows us to study dynamic processes in single crystals of biomolecules.”
ASU’s contribution to the new study involved a combined effort of faculty in the Biodesign Institute’s Center for Applied Structural Discovery, including faculty from the School of Molecular Sciences Petra Fromme and Raimund Fromme and the Department of Physics John Spence and Uwe Weierstall along with their teams of researchers and students, including: research scientist Jay-How Yang , graduate students Shatabdi Roy-Chowdhury, Shibom Basu, Jesse Coe and Chelsie Conrad, undergraduate student Alexander Schaffer from the School of Molecular Sciences, and graduate students Daniel James and Garrett Nelson from the Department of Physics.
In addition to ASU, CFEL and SLAC, the following institutions contributed to the study: University of Hamburg and the Center for Ultrafast Imaging in Germany; the University of Wisconsin, Milwaukee; and the Foundation for Research and Technology-Hellas in Greece. The research was supported by the Helmholtz Association, Germany; the Deutsche Forschungsgemeinschaft, Germany; the European Research Council; the Federal Ministry of Education and Research of Germany; the University of Hamburg, Germany; the BioXFEL Science Technology Center; the U.S. National Institutes of Health, National Institute of General Medical Sciences; and the DOE Office of Science, Office of Basic Energy Sciences.
Contact info:
Petra Fromme
Center for Applied Structural Discovery, The Biodesign Institute, School of Molecular Sciences
Phone: 1 480 965 9028
E-mail: pfromme@asu.edu
More University news

Provost Teaching Awardees, Charter Professors empower local communities, students
The ASU Charter embodies the university’s commitment to student success and research of public value and responsibility to…

New online Bachelor of Social Work program exceeding enrollment expectations
Social workers are in big demand.Citing U.S. Bureau of Labor Statistics figures, the National Association of Social Workers…

Miki Kittilson appointed dean of ASU's College of Global Futures
Miki Kittilson has been appointed dean of Arizona State University’s College of Global Futures, a unit of the Julie Ann…