Study: Combining info on genes, gut bacteria enhances early disease detection
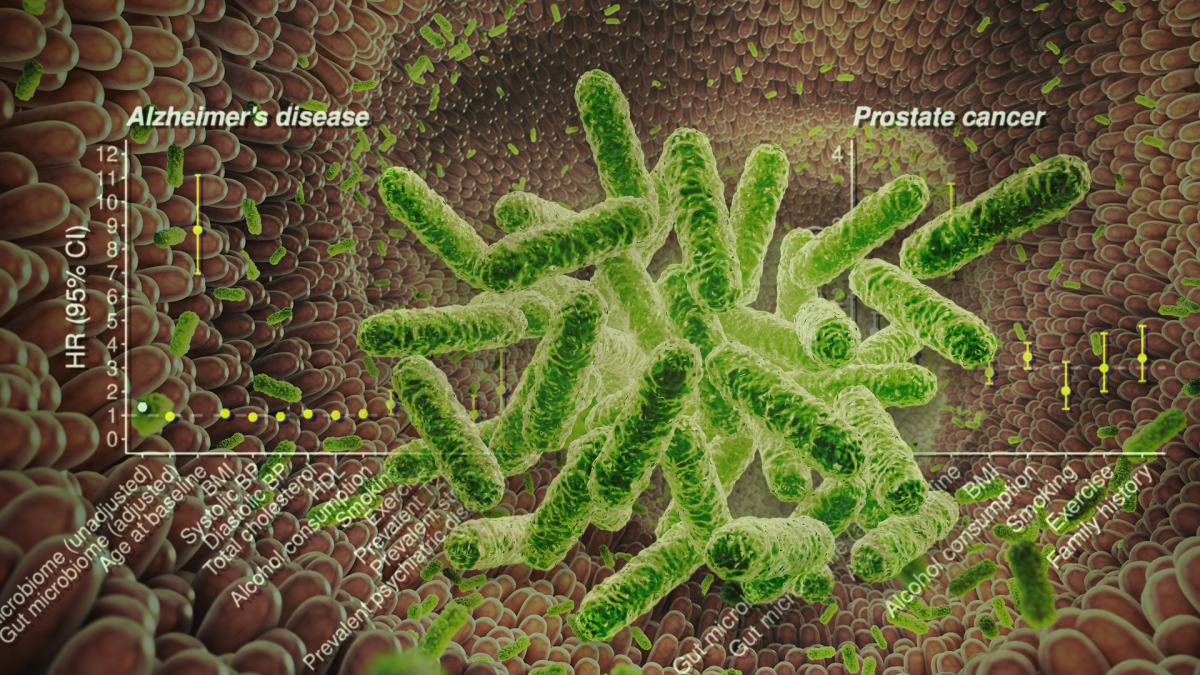
A new study highlights improvements in estimating the risk of common diseases when data from the body's gut microbiome is combined with genetic and other risk factors. Graphic by Jason Drees
Identifying those at highest risk for developing common chronic diseases like heart disease, diabetes, Alzheimer’s disease and cancer is a core priority for preventive medicine. By catching elevated risk early, physicians can implement lifestyle interventions, closer monitoring and targeted therapies to potentially delay or even prevent disease onset and progression.
Traditional risk assessment models based solely on factors like age, family history and standard clinical measurements often fail to capture the full scope of an individual's inherited and biological susceptibility.
A new study demonstrates the power of integrating cutting-edge microbiome analysis into disease-risk prediction models, paving the way for more personalized and effective preventive strategies.
Arizona State University's Qiyun Zhu is part of an international research team, spearheaded by Yang Liu at the University of Cambridge, delving into how microbiomics can refine disease-risk metrics.
Microbiomics examines the complex relationships between a host and its microorganisms, including bacteria, viruses and fungi, to discern their effects on health, disease progression and general wellness.
Zhu is a researcher with the Biodesign Center for Fundamental and Applied Microbiomics and an assistant professor with ASU’s School of Life Sciences.
The study was a collaborative effort involving experts from institutions worldwide, including the University of Cambridge, Baker Heart and Diabetes Institute, Finnish Institute for Health and Welfare, and University of California San Diego. This diverse team combined their expertise in genomics, microbiomics, epidemiology and computational biology to drive the innovative research forward.
The study appears in the current issue of the journal Nature Aging.
Diagnostic challenges
Early detection of many diseases remains crucial for successful therapy. Despite significant advances in disease discovery, early diagnosis for many common ailments remains an inexact science. But research into the body’s vast composition of gut microbial cultures is offering new insights into disease susceptibility.
The gut microbiome serves as an enormous reservoir of microbial life. These complex communities of microorganisms can act synergistically, performing a variety of tasks ranging from digestive activities and weight management to changes in the brain believed to influence behavior.
By combining three complementary components — clinical risk factors, polygenic risk score (a number that indicates a person's inherited risk for developing a particular medical condition) and gut microbiome analysis — the researchers were able to develop integrated predictive models that outperformed any single component alone in forecasting disease risk over long-term follow-up.
The new study gathers health data from more than 5,670 adults, conducted in collaboration by the Baker Heart and Diabetes Institute, the University of Cambridge and the Finnish Institute of Health and Welfare.
Conventional disease-risk assessment typically relies on factors like age, sex, body mass index, blood pressure, cholesterol levels and blood glucose measurements. By leveraging powerful new data streams combining polygenic risk scores with analysis of gut microbial composition, predictive models are enhanced, improving physicians' ability to pinpoint those at elevated risk and tailor prevention strategies to their unique biological profile.
In this study, researchers investigated the individual and combined predictive performance of polygenic risk scores, the gut microbiome and traditional risk factors for development of future coronary artery disease, Type 2 diabetes, Alzheimer's disease and prostate cancer over a median of ~18 years of follow-up using a population survey known as FINRISK.
Multi-pronged analysis
Multi-omics refers to the integration and analysis of data from multiple fields to gain a more comprehensive understanding of biological systems and processes. This approach aims to provide a holistic view of how genetic, transcriptomic, proteomic and metabolomic levels interact within an organism, offering insights into complex biological questions and diseases that cannot be understood through single-omics studies alone.
Multi-omic technologies have uncovered new biomarkers for a variety of common age-related diseases. If individuals at elevated risk for these prevalent chronic illnesses can be flagged early through predictive modeling that incorporates multi-omic biomarkers, preventive interventions could then be applied to high-risk groups before disease onset or progression.
Early preventive strategies like lifestyle modifications, increased monitoring, preventive medications or therapies targeting specific biological mechanisms could potentially delay or even avoid the manifestation of disease symptoms and complications.
As multi-omic technologies continue advancing, their integration into robust predictive models holds promise for ushering in an era of precision prevention where personalized preventive plans can be tailored based on each individual's unique biomolecular profile and risk landscape.
"Multiomics combined with machine learning is revolutionizing how we diagnose diseases and predict disease risks,” Zhu says. “Microbiomics is one critical ‘omics’ among these.”
The new study demonstrates the potential of integrating diverse multi-omic data, including gut microbial composition, to advance the understanding of the development and prediction of diseases. Challenges remain in properly incorporating the diverse data to provide the best risk analyses and improve treatment options.
More Science and technology
ASU startup Crystal Sonic wins Natcast pitch competition
Crystal Sonic, an Arizona State University startup, won first place and $25,000 at the 2024 Natcast Startup Pitch Competition at the National Semiconductor Technology Center Symposium, or NSTC…

Celebrating '20 Years of Discovery' at ASU’s Biodesign Institute
Editor’s note:The Biodesign Institute at Arizona State University continues to celebrate its landmark 20th anniversary with this fifth installment in its "20 Years of Discovery" series. Each…

Rocket science: Students land opportunity to create inflatable lunar pad for NASA
Editor's note: The ASU AEGIS team won the Best Systems Engineering award — one of two awards given to the six teams that presented at the Las Vegas forum held Nov. 11–12. Sixteen Arizona State…
