Cultivating CRISPR innovations

Unlocking the powers of the gene-editing tool called CRISPR is expected to lead to major steps in working with DNA in the human genome to significantly advance medical care. Graphic by Rhonda Hitchcock-Mast/ASU
Few discoveries in science have had the potential for medical impacts as dramatic as the gene-editing tool called CRISPR. Only about a decade after the discovery of the technique, it is being seen as a basis for developing treatments and cures for many diseases and other serious health problems.
An acronym for clustered regularly interspaced short palindromic repeats, CRISPR’s abilities can be used to alter the genomes — the entire set of DNA instructions — in living organisms.
By repairing, disabling or otherwise modifying various genes, the technique has already been used to successfully treat diseases in animals, fight off viruses and prepare pig organs for use in transplants in humans, to name a few of its uses.
Researchers are also exploring multiple possibilities for the use of CRISPR to advance human health care, including refining of CRISPR methodologies and expanding its applications.
Recently, CRISPR has been used also to detect specific nucleic acids, which can enable the high-throughput simultaneous detection of many viruses. For example, by tweaking CRISPR, researchers have designed new rapid diagnostics for SARS-CoV-2, the virus responsible for the COVID-19 pandemic.
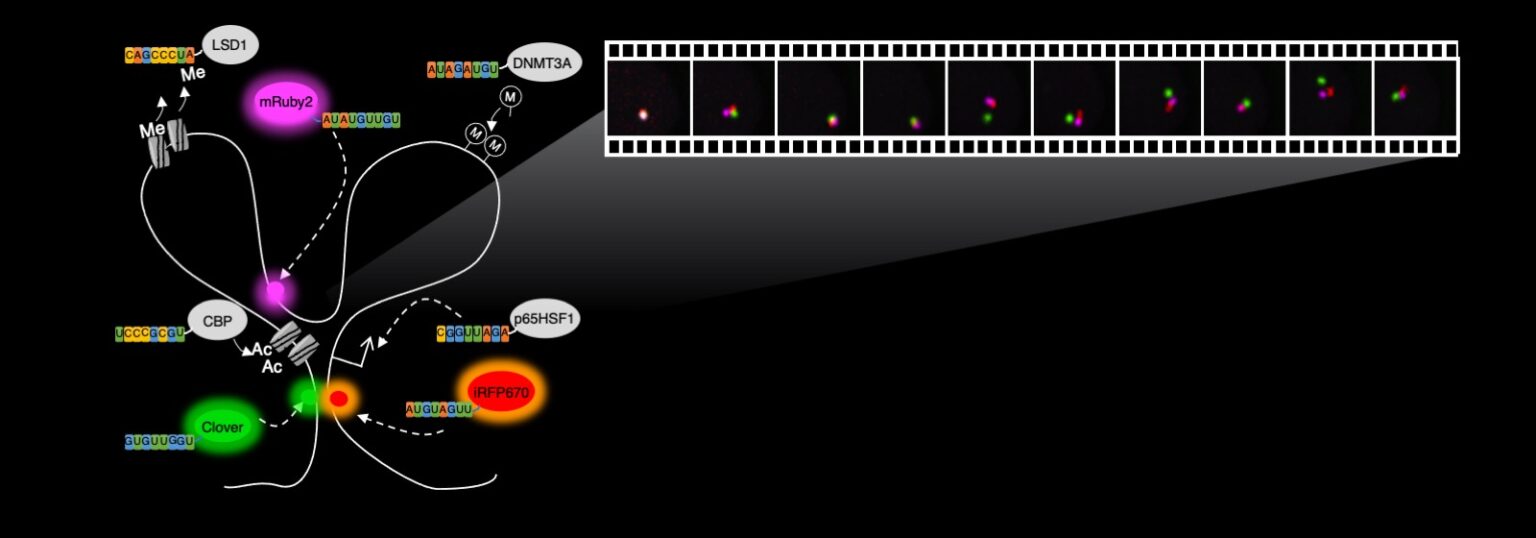
CRISPR can also be engineered to visualize DNA and RNA molecules in living cells, allowing scientists to study the human genome and its regulation as biological processes are taking place in cells. This process, called live-cell genome imaging, also makes it possible to track these molecules as diseases emerge and progress.
A breakthrough in CRISPR-based imaging methods is detailed in a recently published paper in the research journal Nature Communications and a paper in the CRISPR Journal.
Both papers report on research conducted in the Cheng Lab and led by principal investigator Albert Cheng. The headquarters of the project relocated in January of this year when Cheng moved from the Jackson Laboratory for Genomic Medicine to the School of Biological and Health Systems Engineering, one of the seven Ira A. Fulton Schools of Engineering at Arizona State University, where he is now an associate professor.
The Nature Communications paper describes a breakthrough in live-cell genome imaging that allows for the visualization of nonrepetitive genomic sequences with only one gRNA — an unsolved challenge for CRISPR-based imaging since the publication in 2013 of the first CRISPR imaging paper.
One aspect of CRISPR research conducted in the Cheng Lab, depicted in the illustration above, involves the use of fluorescent proteins, or proteins that can emit light in different colors, that are placed in various locations in the genome within the body’s cells. Through the use of fluorescent microscopy, the illuminated genomic locations appear as dots that move over time. The short movie below records how the genome moves. The rapid movements revealed by this method demonstrate that genome folding is a highly active and dynamic process, differing from what had earlier been thought about the process. ASU Associate Professor Albert Cheng says these new insights have implications for the stochasticity — or randomness — and robustness of gene regulatory processes. Image courtesy of the Cheng Lab
Video courtesy of the Cheng Lab
Visualizing gene dynamics in living cells
This new imaging method means that instead of being restricted to clustered repetitive sequences, a majority of the genome can be imaged, enabling a better understanding of the dynamics of genome folding in living cells, Cheng says.
Long genomes must be folded extensively to fit into the three-dimensional space of a cell’s nucleas, which is about 10 nanometers in diameter, and the folding itself is dynamically changing every second to position different locations of the genome in proximity. This enables the right genes and proteins to be expressed at the right time for cells to function properly.
“The CRISPR imaging tool will help us visualize the folding process as it happens in living cells, allowing us to decipher the mysteries of gene regulation,” Cheng says.
The CRISPR Journal paper reports on work to develop computational tools and databases that can be used in the design of probes, called guide RNA, or gRNA, that can be used with the imaging technology described in the Nature Communications paper.
The imaging method has already been put to use in cancer research. The Cheng Lab, in collaboration with colleagues at the Jackson Laboratory, recently published a paper in the journal Cancer Discovery showing how the imaging method was used to track, in living cancer cells, extrachromosomal DNA (ecDNA) — DNA molecules that have “escaped” from the chromosomes and multiplied to tens to hundreds of copies to give cancer cells a growth advantage.
“The understanding of how these ecDNA molecules multiply in cells will also help us better understand how cancer cells grow and devise therapeutics directed toward them,” Cheng says.
He is collaborating on these efforts with Jackie Jufen Zhu, a cancer genomics and imaging expert who also came from the Jackson Laboratory earlier this year and is now an assistant research professor working in Cheng’s lab.
Zhu develops and applies sequencing and imaging methods to study genome folding and transcription factors, especially in the context of cancer biology, with the goal of understanding disease mechanisms and developing targeted therapies.
Research success would enhance medical care capabilities
In addition to developing new genome imaging approaches and applying them to study diseases described in these papers, the Cheng Lab develops and applies technology based on engineered DNA and RNA binding proteins — a process that could enable the regulation of genes to create new therapeutics.
Other directions the lab’s research is taking include making genome editing more precise, developing novel approaches for epigenetic and transcript editing, and developing DNA and RNA sensors and imaging techniques.
Albert Cheng
Success in these pursuits could lead to meaningful leaps forward in the fields of genomics, epigenetics, gene regulation, stem cell biology and cancer treatment.
“We want to more accurately decipher the gene regulation mechanisms that underlie the development of many diseases to help us pinpoint new therapeutic targets,” Cheng says. “We hope that by better understanding the mechanisms of gene regulation and creating new gene therapies, it will lead to both more effective treatment options for diseases like cancer, and especially for diseases for which there are currently no available treatments.”
Cheng and his colleagues are already using epigenetic editing techniques that “turn on” specific genes that can specifically kill cancer cells.
Other researchers have applied Cheng’s epigenetic editing tools to increase insulin production in cells or to convert liver cells into islet-like cells, which may be potential treatment strategies for diabetes. In the future, Cheng hopes to combine molecular parts that he and others have built to engineer “smart therapeutics” that can respond to cellular condition and provide therapeutic effects “on demand.”
Cheng will begin passing on his growing knowledge of CRISPR technology to ASU students when he begins teaching the biomedical engineering course BME 362 Methods in Molecular Cellular Biology in fall 2022.
More Science and technology

ASU and Deca Technologies selected to lead $100M SHIELD USA project to strengthen U.S. semiconductor packaging capabilities
The National Institute of Standards and Technology — part of the U.S. Department of Commerce — announced today that it plans to…
From food crops to cancer clinics: Lessons in extermination resistance
Just as crop-devouring insects evolve to resist pesticides, cancer cells can increase their lethality by developing resistance to…

ASU professor wins NIH Director’s New Innovator Award for research linking gene function to brain structure
Life experiences alter us in many ways, including how we act and our mental and physical health. What we go through can even…