New research in protein sequencing poised to transform medicine
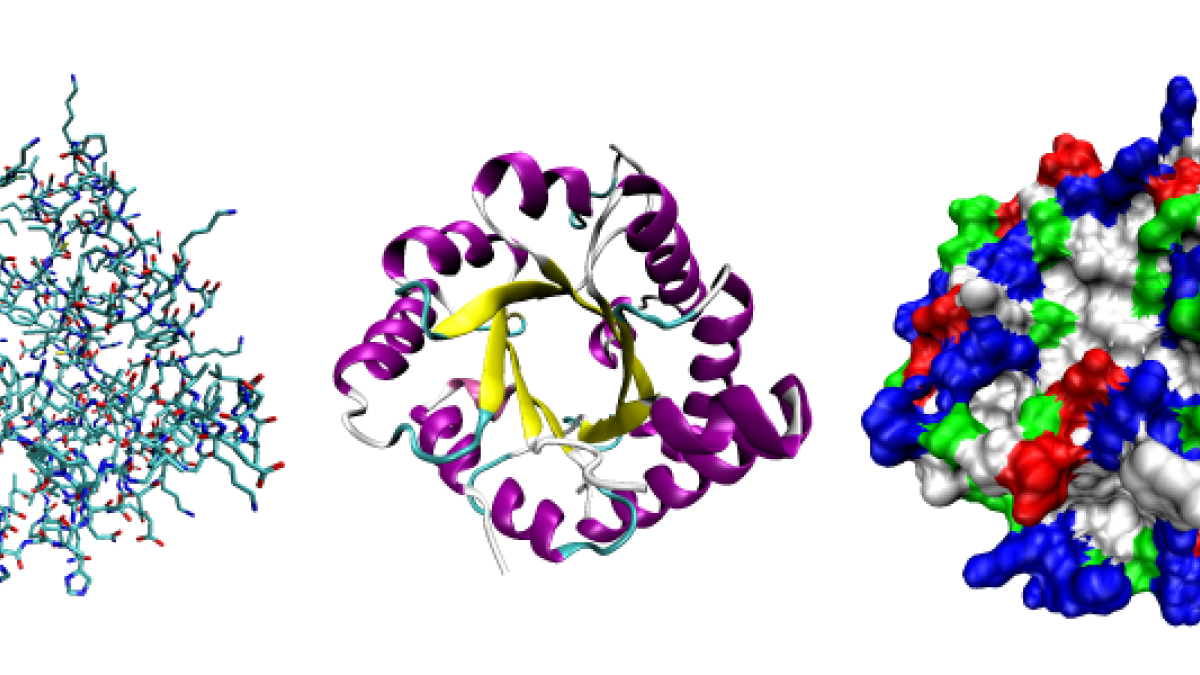
Proteins are composed of 20 amino acids that fold into elaborate, three-dimensional forms. Seen here, three possible representations of the three-dimensional structure of the protein triose-phosphate isomerase. Graphic courtesy Opabinia regalis.
While DNA provides the genetic recipe book for biological form and function, it is the job of the body’s proteins to carry out the complex commands dictated by DNA’s genetic code.
Stuart Lindsay, a researcher at the Biodesign Institute at Arizona State University, has been at the forefront of efforts to improve rapid DNA sequencing and has more recently applied his talents to explore the much thornier problem of sequencing protein molecules, one molecule at a time.
A new overview article describes Lindsay’s efforts, along with those of international colleagues, toward applying a variety of innovative strategies for protein sequencing at the single-cell, and even single-molecule, level.
”One of the surprises of the Human Genome Project was that humans can be built from a mere 25,000 genes," Lindsay said. "The real complexity turns out to lie in the almost infinite number of ways proteins can be put together from this limited set of genes, and in the ways individual amino acids — the building blocks of proteins — can be chemically modified. For these reasons we need tools to analyze proteins at the single-molecule level.”
Lindsay is the director of Biodesign’s Center for Single-Molecule Biophysics, the Edward and Nadine Carson Presidential Chair in Physics at ASU and Regents Professor in The College of Liberal Arts and Sciences in chemistry and biochemistry.
Stuart Lindsay is the director of Biodesign’s Center for Single-Molecule Biophysics, the Edward and Nadine Carson Presidential Chair in Physics at ASU and Regents Professor in The College of Liberal Arts and Sciences.
The review was jointly supervised by Javier Alfaro, University of Gdańsk, Poland; Amit Meller, Technion–Israel Institute of Technology, Haifa, Israel; Chirlmin Joo, Delft University of Technology, the Netherlands; and Adam Pomorski, University of Wroclaw, Poland.
The research appears in the current issue of the journal Nature Methods.
Although cells may superficially appear identical, they are in reality unique entities with individual properties that may differ dramatically, with important implications for health and disease. Since proteins do most of the work within the cell, investigating cell-to-cell variation in organs, tissues and cell cultures can provide critical insights, particularly for the study of cancer, where individual cell mutations can become amplified, driving the progress of the disease.
The field of protein research (known as proteomics) has traditionally been dominated by mass-spectrometry technologies, which are expensive and generally only sequence batch proteins contained in complex mixtures. Ferreting out single proteins in individual cells will require a transformation in sequencing technology similar to the DNA sequencing revolution. According to Alfaro, “the problem is that unlike DNA, there is no way to amplify proteins and this poses a big challenge to the sensitivity of current instrumentation."
Lindsay’s approach to the problem involves a technique known as recognition tunneling, which relies on a technology known as scanning tunneling microscopy (STM). In earlier research, Lindsay applied STM to identify DNA bases sequentially fed through a tiny aperture known as a nanopore, much as one might feed thread through the eye of a needle.
As DNA nucleotides transit through the nanopore, they are momentarily held captive between a pair of electrodes. Current passing through each base, known as tunneling current, produces tell-tale electrical spikes, which are like a fingerprint of each of the four DNA bases: adenine, thymine, cytosine and guanine.
Recent advances have allowed the same basic idea to be applied to the identification of proteins, through sequential identification of the 20 amino acids from which all proteins are built. While recognition tunneling of proteins follows the same basic game plan used to sequence DNA bases, the task is far more delicate for protein sequencing, requiring the bonding of amino acids to specialized adapter molecules for the tunneling current to pass through as well as machine learning algorithms to help pick out the precise amino acid being sequenced.
”This review is part of the launch of a worldwide effort to make single-molecule protein sequencing a reality, with many labs around the world contributing to a variety of new technologies, our electronic approach being one,” Lindsay said.
“We expect that these groundbreaking technologies will penetrate into the market, particularly in pharmaceutical industry, academic research and clinical diagnostics,” Joo said. “Using these technologies, we will be able to uncover the diversity of the proteome, study how the proteins communicate and interact, and provide new insights into how life actually works at the molecular level.”
Continued advancement in protein sequencing may open the door to routine clinical application for diagnostic and treatment purposes, fulfilling the longstanding ambition of personalized medicine.
More Science and technology

Compact X-ray laser lab aims to reveal deep secrets of life, matter and energy
X-rays allow us to view inside the human body to diagnose broken bones and other hidden problems. More recent X-ray advances are…

Apollo lunar samples enable ASU researcher to pinpoint moon’s crystallization timeline
A team of researchers, including Arizona State University geochemist Melanie Barboni, in collaboration with scientists from The…

NASA launches space telescope to chart the sky and millions of galaxies
California’s Vandenberg Space Force Base was the site for Tuesday’s 8:10 p.m. launch of the NASA SPHEREx mission aboard a SpaceX…


