ASU research plays key role in lower Salt River restoration

The Salt River in Arizona. Photo courtesy of Depositphotos.
Along the lower banks of the Salt River, dense thickets of tall, weedlike invasive saltcedar and giant reed threaten native vegetation vital to preserving the biodiversity of one of the few remaining low elevation desert riparian vegetation communities in the state.
Accurately mapping species level vegetation is an essential step in managing invasion risk, guiding remediation efforts and intervention strategies and ultimately understanding what processes are facilitating invasive species growth or expansion.
However, mapping large areas of vegetation manually from the ground is both labor- and time-intensive, and using traditional satellite platform imagery is difficult due to its coarse spatial resolution. Unmanned aircraft systems (UAS), or drones, provide a largely new opportunity for high-resolution remote sensing and capturing this critical data.
Recently published research led by Arizona State University Master of Advanced Study in Geographic Information Systems (MAS-GIS) alumnus Arnold Chi Kedia and co-authored by Amy Frazier, assistant professor in the School of Geographical Sciences and Urban Planning, uses drone-sensed data to spatially map vegetation along the lower Salt River and distinguish native and non-native vegetation species.
In their paper, “An Integrated Spectral-Structural Workflow for Invasive Vegetation Mapping in an Arid Region Using Drones,” published in Drones, the researchers develop a new mapping process combining structural data with spectral data to three-dimensionally model the lower Salt River landscape with precise resolution within 3-5 centimeters.
“Not only do we have the spectral reflectance, how different vegetation species are reflecting light, but we also have their structure in terms of how tall they are or how voluminous they are,” said Frazier, who also is the associate director of ASU’s Geospatial Research and Solutions. “Both of those data streams came from drone data, and the combination of adding the structure actually made our accuracy for figuring out where things were much higher.”
The researchers found that including spectral and three-dimensional structural layers in their classification, as opposed to just using spectral data, improved overall vegetation species identification accuracy from 80% to 93%.
“Using terrain data and canopy height were actually very important characteristics to identify these plant species,” Kedia said. “Most research just uses spectral data, but structural data controls how plant species are distributed in their natural habitat. This is what makes our paper unique; this method of combining both structural and spectral information has not been used before.”
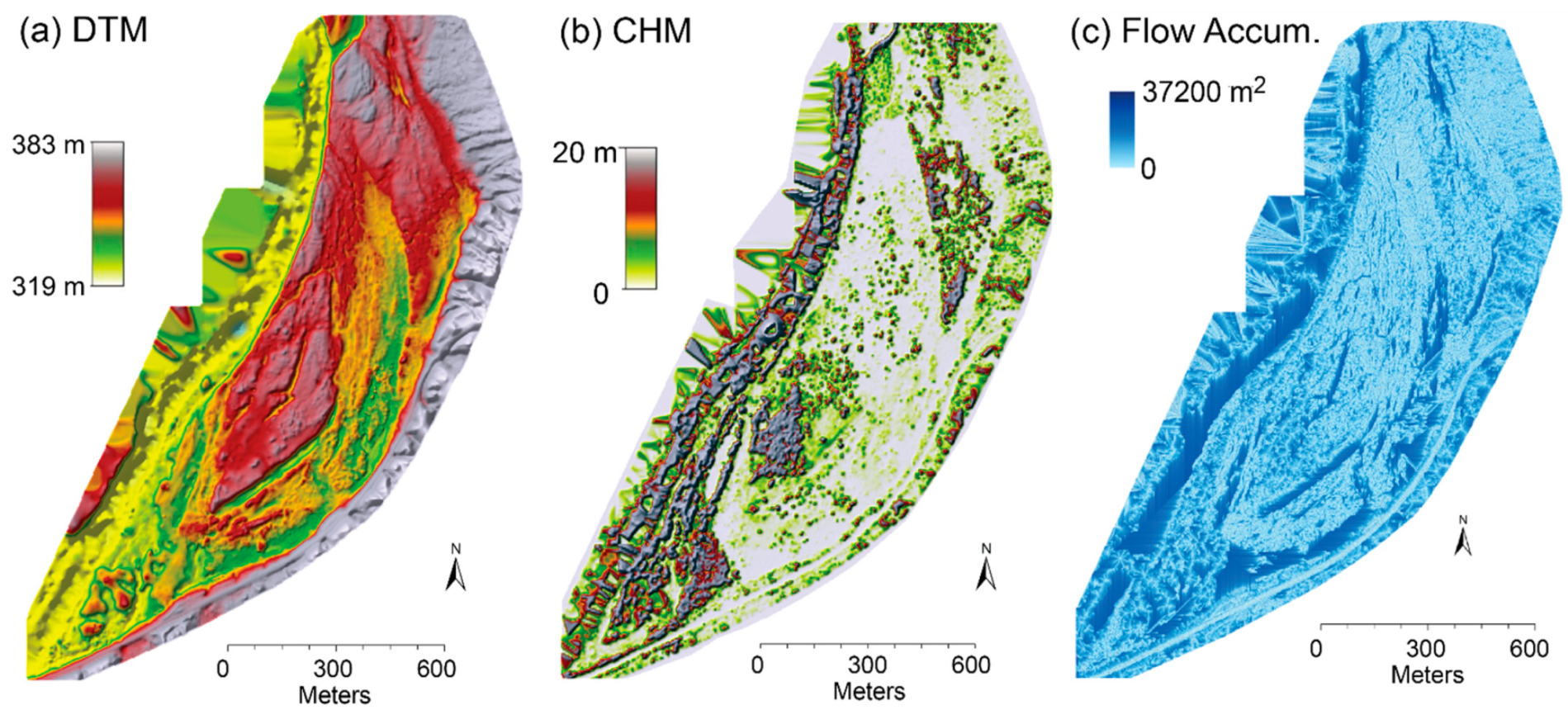
The three structural layers, including (a) digital terrain model (DTM), (b) canopy height model (CHM) and (c) flow accumulation.
With a more accurate and detailed picture of what’s happening, land managers can use the information to better develop strategies to monitor and control invasive vegetation in the lower Salt River.
The research was conducted in collaboration with local drone company Green Drone AZ, and the data from the study is now being used in the continuing Lower Salt River Restoration Project.
Additional co-authors of the research include ASU MAS-GIS graduates Brandi Kapos and Songmei Liao, and Green Drone AZ team members Jacob Draper, Justin Eddinger and Christopher Updike.

