Massive simulation reveals how a bacterial organelle converts sunlight to chemical energy

Researchers used supercomputers to construct a 136 million-atom model of the chromatophore, a primitive light-harvesting structure in purple bacteria. The simulated organelle behaved just as it does in nature, the team reports. Graphic by Christopher Maffeo
Researchers from ASU’s Biodesign Institute, in collaboration with colleagues from the University of Illinois, report that they have successfully simulated every atom of a light-harvesting structure in a photosynthetic bacterium that generates energy for the organism. The simulated organelle behaves just like its counterpart in nature, the researchers report.
The work is a major step toward understanding how some biological structures convert sunlight into chemical energy, a biological innovation that is essential to life, the researchers said. They report their findings in the journal Cell.
The research team, originally led by University of Illinois physics professor Klaus Schulten, continued the work after Schulten’s death in 2016. The study fulfills, in part, Schulten’s decades-long dream of discovering the mechanisms by which atomic-level interactions build and animate living systems.
Schulten decided very early in his career to study photosynthetic systems. Schulten and research scientist Melih Sener, a co-author, have modeled the chromatophore, a photosynthetic organelle that produces chemical energy in the form of a molecule known as ATP, with the help of experimental data from Neil Hunter from the University of Sheffield in a decade-long collaboration. Schulten was fascinated with the chromatophore from purple bacteria as one of the most primitive biological light-harvesting apparatus known.
“He was a physicist; he wanted to understand biology at the physics level,” said Illinois biochemistry professor Emad Tajkhorshid, a co-author on the new study. “But then he realized biology only works if you put all of the complexity into the model. And the only way to do that was with supercomputers.”
Over the years, Schulten recruited and supported collaborators at Illinois and elsewhere to help him tackle the challenge. The team constructed a 136 million-atom hybrid model of the chromatophore using structural and spectroscopic data derived by long-term collaborator Hunter. The effort required a colossal amount of supercomputer power over a period of four years. The work was conducted on the Titan and Summit supercomputer at the Oak Ridge National Laboratory in Knoxville, Tennessee, and on Blue Waters, which is housed at the National Center for Supercomputing Applications at the University of Illinois.
Schulten and his colleagues had already conducted molecular simulations of many of the individual protein and lipid components of the chromatophore, which produces the ATP needed to power a living cell.
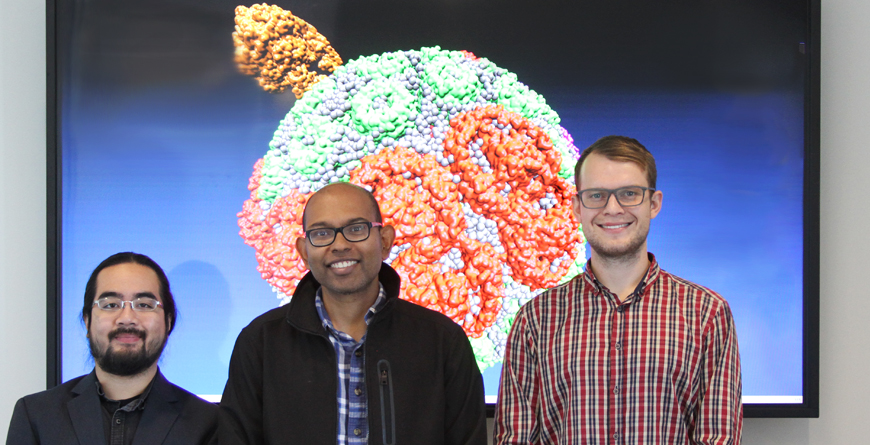
“The chromatophore has an antenna, a battery and a motor,” said the article's lead author, Abhishek Singharoy, who worked with Schulten at Illinois as a Beckman Fellow and CPLC Fellow before moving to Arizona State University in Tempe in December 2017. The antenna harvests light, the battery directs that energy to the motor and the motor cranks out ATP, said Singharoy, who is now a researcher in the Biodesign Center for Applied Structural Discovery and professor in the School of Molecular Sciences at ASU. John Vant and Jonathan Nguyen, fellow researchers in the Singharoy lab, also participated in the new study. Singharoy has focused on state-of-the-art computational approaches for capturing cell-scale biological responses with atomic precision. For the current study, his group was able to parse the data from ORNL’s massive Summit supercomputer using the Agave cluster, a high-performance computing resource at ASU.
ASU research team (from left): Jonathan Nguyen, Abhishek Singharoy and John W. Vant, framed by a graphic of the chromatophore molecule they modeled using supercomputing resources. Photo by Shireen Dooling
Figuring out how the system worked required putting all the parts together, said Illinois physics professor Aleksei Aksimentiev, who guided the project to completion after Schulten’s death. This meant dissecting the chromatophore with every tool available to science, from laboratory experiments to electron microscopy, to programming innovations that broke down the computing challenge into manageable steps, Aksimentiev said.
Once they had a working model of the chromatophore, the researchers watched simulations that revealed how the organelle functioned under different scenarios. They changed the concentration of salt in its environment, for example, to see how it coped with stress.
When they exposed their simulated organelle to conditions that it typically experiences in the cell, they were surprised by how it behaved. It immediately became less spherical, and certain proteins embedded in the membrane began to clump together.
“We started with a perfect sphere, but very rapidly it became imperfect, with flat areas and little areas with high curvatures,” Aksimentiev said. “And all of that, our calculations reveal, has a biological role.”
The protein arrangement creates patches of positive and negative charges that facilitate the distribution of electrons across the system, the researchers said. The electrons are ultimately swapped for protons that drive an enzyme known as an ATP synthase, the motor that produces ATP.
“Chromatophore structure is like a circuit diagram,” said study co-author Melih Sener, a research scientist at the Beckman Institute for Advanced Science and Technology, where much of the computational work was conducted. “If you know how energy and charges travel in it, you know how this machine works. Chromatophore is basically an electronic device.”
The study confirms that, at the atomic scale, physics drives biology, the researchers said. The work will inform future studies of more complex energy-generating organelles in other microorganisms, and in plants and animals, they said. And it will advance scientists’ understanding of nature’s solution to a perpetual human problem: how to efficiently extract energy from one’s environment without poisoning oneself.
The research was made possible through the National Institutes of Health Center for Macromolecular Modeling and Bioinformatics and the National Science Foundation Center for the Physics of Living Cells, both at the University of Illinois. Aksimentiev and Tajkhorshid also are also affiliates of the Beckman Institute. All the computations were supported by first Schulten’s and then Singharoy’s Innovative and Novel Computational Impact on Theory and Experiment (INCITE) grant with the Department of Energy. The Arizona leg of the work in Singharoy’s laboratory was also supported by Research Corporation for Science Advancement and the Gordon and Betty Moore Foundation.
Grants: P41-GM104601, NIH R01-GM067887, NSF PHY-1430124, MCB1616790,
Written by Diana Yates and Richard Harth
More Science and technology

Phoenix-based startup taps ASU alum to develop digital coach to help with menopause
It started with secret conversations behind the scenes at technology conferences.But Susan Sly and her colleagues weren’t…

Damaged moon rocks persuade researchers to rethink lunar history
New research reveals that even lunar samples once believed to be pristine have been extensively altered by meteorite impacts,…

CSI: Mars
By Wendee NicoleTravelling to Mars has loomed large in the public’s imagination since at least the late 1800s when H.G. Wells' “…