Electrifying science: New study describes conduction through proteins
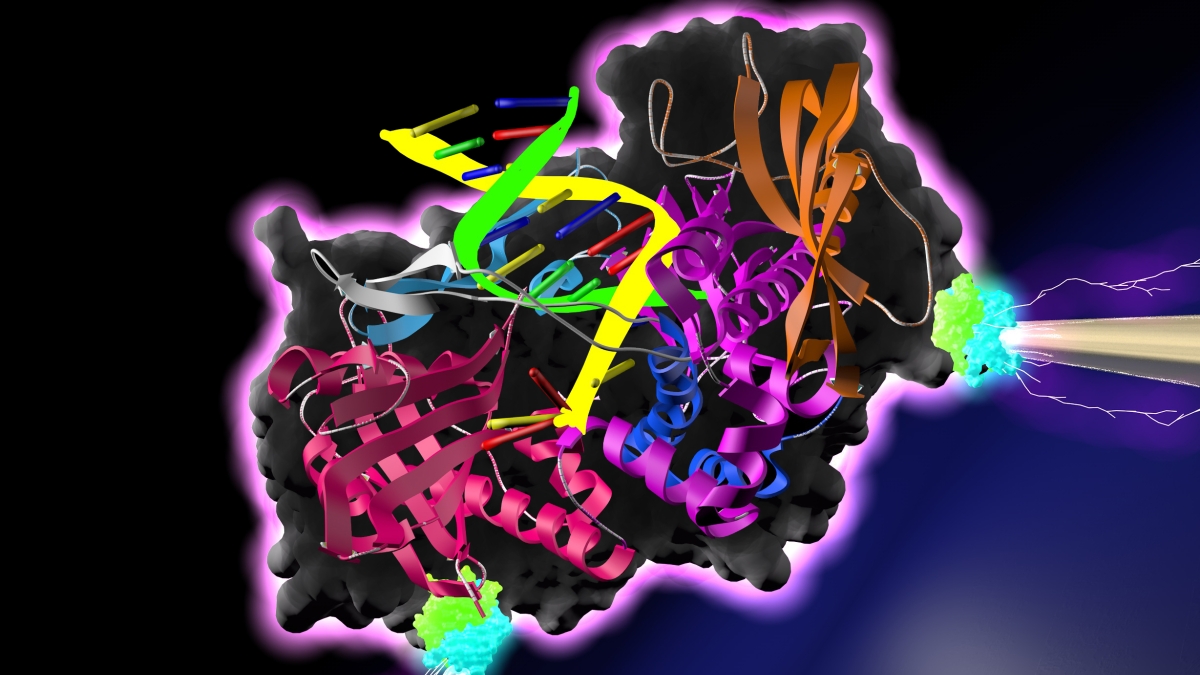
A DNA polymerase — an enzyme that synthesizes DNA molecules from nucleotide building blocks — is poised between a pair of electrodes. Binding of nucleotides by the polymerase causes conductance spikes, which may be characteristic of the specific molecules bound by the polymerase. Such a device, in theory, could carry out rapid, accurate, low cost DNA sequencing, and may have many additional applications in medical diagnostics, industrial production and other areas.
Amid the zoo of biomolecules essential to life, enzymes are among the most vital. Without these specialized proteins, which speed up the rates of chemical reactions, thousands of essential life processes, from cell growth and digestion to respiration and nerve function, would be impossible.
In new research, Arizona State University Professor Stuart Lindsay and his colleagues investigate a recently discovered feat carried out by enzymes and, most likely, all proteins. Under proper conditions, they can act as superb conductors of electricity, permitting them to be incorporated into a range of electronic devices.
“It is a way of plugging the amazing chemical diversity of enzymes directly into a computer,” Lindsay said.
While the role of protein conductance in nature remains a matter of mystery and speculation, harnessing this phenomenon for human use will likely open new avenues for biochemical sensing devices, smart industrial production and new innovations in medical diagnostics.
Perhaps most exciting, electrical conductance through a special type of enzyme may signal a significant advance for DNA sequencing. Using a DNA polymerase, nature’s own high-resolution DNA reader, in such a device could potentially allow for lightning-fast sequencing of entire human genomes with unprecedented accuracy at very low cost. The new study “opens the Pandora's box of looking at the function of any enzyme in a computer chip.”
Current affairs
Authors of the new study describe the tricks they used to affix a DNA polymerase to a pair of electrodes and the resulting current spikes associated with the enzyme successively binding and releasing target DNA nucleotides. The successful demonstration of enzyme conductance paves the way for eventually mounting arrays of proteins onto computer chips, where they can act as biological parallel processors for a variety of tasks.
“Enzymes are incredible molecules that carry out chemical reactions that just wouldn't happen otherwise,” Lindsay said. To give a sense of the power of these molecules, certain reactions essential to life processes, unfolding thousands of times per second, would require millennia to occur in the absence of enzymes.
Lindsay directs the Biodesign Center for Single Molecule Biophysics at ASU. The center’s primary research focuses on science at the nexus of molecular medicine and nanotechnology.
His group’s findings appear in the forthcoming edition of the journal ACS Nano.
Stuart Lindsay directs the Biodesign Center for Single Molecule Biophysics. He is also a Regents Professor and Nadine and Edward Carson Professor of Physics and Chemistry.
Proteins as conductors
Until quite recently, proteins were regarded strictly as insulators of electrical current flow. Now, it seems, their unusual physical properties may lead to a condition in which they are sensitively poised between an insulator and a conductor. (A phenomenon known as quantum criticality may be at the heart of their peculiar behavior.)
Indeed, in earlier research, Lindsay demonstrated strong electrical conductance through a protein captured between a pair of electrodes. The new research carries the investigations of protein conductance a step further. Previously, the protein was hooked up via its two so-called active sites. These are the regions of a protein that bind selected molecules, often resulting in a conformational change in the molecule’s complex 3D structure and the completion of the protein’s given task.
This time, the biomolecule was sensitively wired to the electrodes by means of alternate binding sites on the enzyme, leaving the active sites available to bind molecules and carry out natural protein function.
Nature’s Kindle
The enzyme molecule chosen for the experiments is one of the most important for life. Known as a DNA polymerase, this enzyme binds with successive nucleotides in a length of DNA and generates a complimentary chain of nucleotides, one by one. This versatile nanomachine is used in living systems for copying DNA during cell replication as well as for repairing breaks or other insults to the DNA.
The study describes techniques for affixing the DNA polymerase to electrodes so as to generate strong conductance signals by means of two specialized binding chemicals known as biotin and streptavidin. When one electrode was functionalized using this technique, small conductance spikes were generated as the DNA polymerase successively bound and released each nucleotide, like a grasping hand catching and releasing a baseball. When both electrodes were outfitted with streptavidin and biotin, much stronger conductance signals, measuring three to five times as large, were observed.
The idea for leveraging a polymerase to carry out rapid DNA sequencing has been with Lindsay for awhile. He had considered using it in earlier devices he created in which sections of DNA were fed through narrow tunnel junctions.
“Wouldn't it be neat if you could put a pair of electrodes inside of polymerases because the polymerase grabs the DNA and chugs it through the junction. If you had a readout mechanism embedded in the polymerase, you've got the ideal sequencing machine,” he said.
The new method hopes to take a different approach, using the polymerase’s own speed-reading expertise to provide a readout of nucleotides through conductance spikes specific to each of the four DNA bases. In practice, a number of design hurdles must be overcome. Proper attachment of the polymerase for electrical conductance is a delicate affair and involved much trial and error. Binding sites must be engineered at specific domains that do not affect protein folding and function and connections had to be devised to prevent the enzyme itself from making contact with the electrodes. The use of biotin for binding the molecule also appears to be critical for high conductance. Biotin binding a pocket of the streptavidin appears to help drive electron transport deep into the protein interior, thereby maximizing conductance.
Separating conductance signals registering each successive DNA base from background noise and random motions of the contact points for the enzyme has also proven challenging and sophisticated machine learning algorithms are being brought to bear to clarify the conductance readouts. Lindsay believes many of these noise issues will be resolved when the polymerases are incorporated into properly isolated and sealed chips holding the enzyme rigidly in place.
Enzyme frontiers
The first complete human genome was a milestone for science and medicine. The herculean effort by the Human Genome Project consumed 13 years of labor at a cost of a billion dollars. Now the floodgates to a new era of protein bioelectronics may be opening, with many surprises likely in store.
If remaining technical hurdles can be overcome, DNA sequencing could be carried out at the breakneck speed of a functional DNA polymerase, or around a hundred nucleotides per second. “If you put 10,000 molecules on a chip — not a hard thing to do — you'll sequence a whole genome in under an hour,” Lindsay said.
More Science and technology
From food crops to cancer clinics: Lessons in extermination resistance
Just as crop-devouring insects evolve to resist pesticides, cancer cells can increase their lethality by developing resistance to…

ASU professor wins NIH Director’s New Innovator Award for research linking gene function to brain structure
Life experiences alter us in many ways, including how we act and our mental and physical health. What we go through can even…

ASU postdoctoral researcher leads initiative to support graduate student mental health
Olivia Davis had firsthand experience with anxiety and OCD before she entered grad school. Then, during the pandemic and as a…
