For the past few decades, some scientists have known the shape of things to come in nanotechnology is tied to the molecule of life, DNA.
This burgeoning field is called "DNA origami." The moniker is borrowed from the art of conjuring up birds, flowers and other shapes by imaginatively folding a single sheet of paper.
Similarly, DNA origami scientists are dreaming up a variety of shapes — at a scale one thousand times smaller than a human hair — that they hope will one day revolutionize computing, electronics and medicine.
Now, a team of Arizona State University and Harvard scientists has invented a major new advance in DNA nanotechnology. Dubbed “single-stranded origami” (ssOrigami), their new strategy uses one long noodle-like strand of DNA, or its chemical cousin RNA, that can self-fold — without even a single knot — into the largest, most complex structures to date.
And the strands forming these structures can be made inside living cells or using enzymes in a test tube, allowing scientists the potential to plug-and-play with new designs and functions for nanomedicine: picture tiny nanobots playing doctor and delivering drugs within cells at the site of injury.
“I think this is an exciting breakthrough, and a great opportunity for synthetic biology as well,” said Hao Yan, a co-inventor of the technology, director of the ASU Biodesign Institute’s Center for Molecular Design and Biomimetics, and the Milton Glick Professor in the School of Molecular Sciences.
“We are always inspired by nature’s designs to make information-carrying molecules that can self-fold into the nanoscale shapes we want to make,” he said.
As proof of concept, they’ve pushed the envelope to make 18 shapes, including emoji-like smiley faces, hearts and triangles, that significantly expand the design studio space and material scalability for so-called, “bottom-up” nanotechnology.
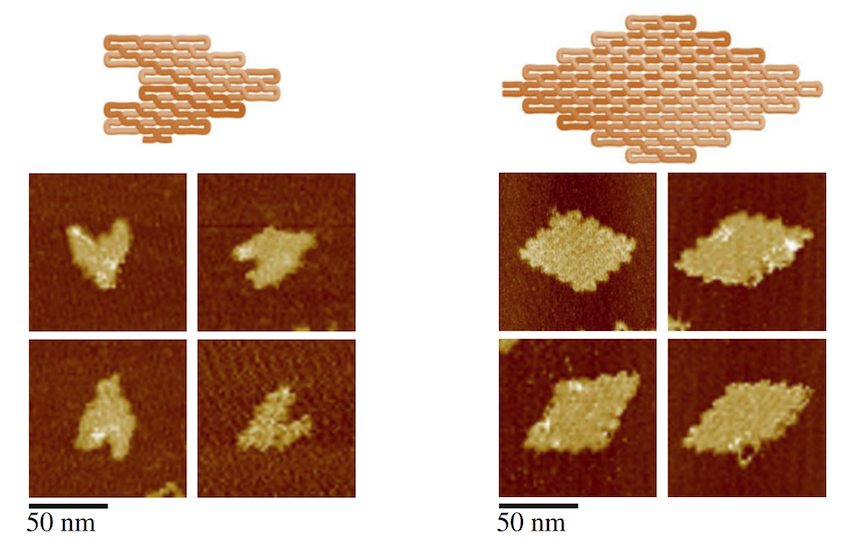
Two DNA origami structures in the shape of a heart and rhombus. Photo courtesy Biodesign Institute
Size matters
To date, DNA nanotechnology scientists have had to rely on two main methods for making spatially addressable structures with finite dimensions.
The first was molecular bricks: small, short pieces of DNA that can fold together to make a single structure. The second method was scaffolded DNA, where a single strand is shaped into a structure using helper strands of DNA that staple the structure into place.
“These two methods are not very scalable in terms of synthesis,” said Fei Zhang, a senior co-author on the paper and Biodesign assistant research professor. “When you have so many short pieces of DNA, you can’t replicate it using biological systems."
Furthermore, each method has been limited because as the size of the structure increases, the ability to fold correctly becomes more challenging.
Now, there is a new way.
For Yan and his team to make their breakthrough, they had to go back to the drawing board, which meant looking at nature for inspiration. They found what they were looking for with a chemical cousin of DNA, in the form of complex RNA structures.
The complex RNA structures discovered to date contain single-stranded RNA molecules that self-fold into structures without any topological knots. Could this trick work for single-stranded DNA or RNA origami?
They were able to crack the code of how RNA makes structures to develop a fully programmable ssOrigami architecture.
“The key innovation of our study is to use DNA and RNA to construct a structurally complex yet knot-free structure that can be folded smoothly from a single strand,” Yan said. "This gave us a design strategy to allow us to fold one long strand into complex architecture.
“With help from a computer scientist in the team, we could also codify the design process as a mathematically rigorous formal algorithm and automate the design by developing a user-friendly software tool.”
The algorithm and software were validated by the automated design and experimental construction of six distinct DNA ssOrigami structures (four rhombuses and two heart shapes).
The goal of Hao Yan's research group is to achieve programmed design and assembly of biologically inspired nanomaterials. Photo by Deanna Dent/ASU Now
Form plus function
It’s one thing to make crafty patterns and smiley faces with DNA, but critics of DNA origami have been wondering about the practical applications.
“I think we are much closer to real practical applications of the technology,” Yan said. “We are actively looking at the first nanomedicine applications with our ssOrigami technology.”
They were also able to demonstrate that a folded ssOrigami structure can be melted and used as a template for amplification by DNA copying enzymes in a test tube and that the ssOrigami strand can be replicated and amplified via clonal production in living cells.
“Single-stranded DNA nanostructures formed via self-folding offer greater potential of being amplifiable, replicable and cloneable, and hence the opportunity for cost-efficient, large-scale production using enzymatic and biological replication, as well as the possibility for using in-vitro evolution to produce sophisticated phenotypes and functionalities,” Yan said.
These same design rules could be used for DNA’s chemical cousin, RNA.
A key design feature of ssOrigami is that the strand can be made and copied in the lab and in living cells and subsequently folded into designer structures by heating and cooling the DNA.
To make it inside the lab, they used the photocopier of cloning sequences, called PCR, to replicate and produce ssDNA.
Inside living cells, they first placed it inside a mule of molecular cloning, called a plasmid, after it was placed into a common lab bacteria called E. coli cells. When they treated the bacteria with enzymes to free up the ssDNA, they could isolate it, and then fold it into its target structure.
“Because plasmid DNA can be easily replicated in E. coli, the production can be scaled up by growing a large volume of E. coli cells with low cost,” Yan said. This gets around the constraint of having to synthesize all of the DNA in the lab from scratch, which is far more expensive.
It also moves them in a direction where they can potentially make the structures inside of cells.
“Here we show bacteria to make the strand, but still need to do thermal annealing outside the bacteria to form the structure,” Yan said. “The ideal situation would be to design an RNA sequence that can get transcribed inside the bacteria, and fold inside the bacteria so we can use bacteria as a nanofactory to produce the material.”
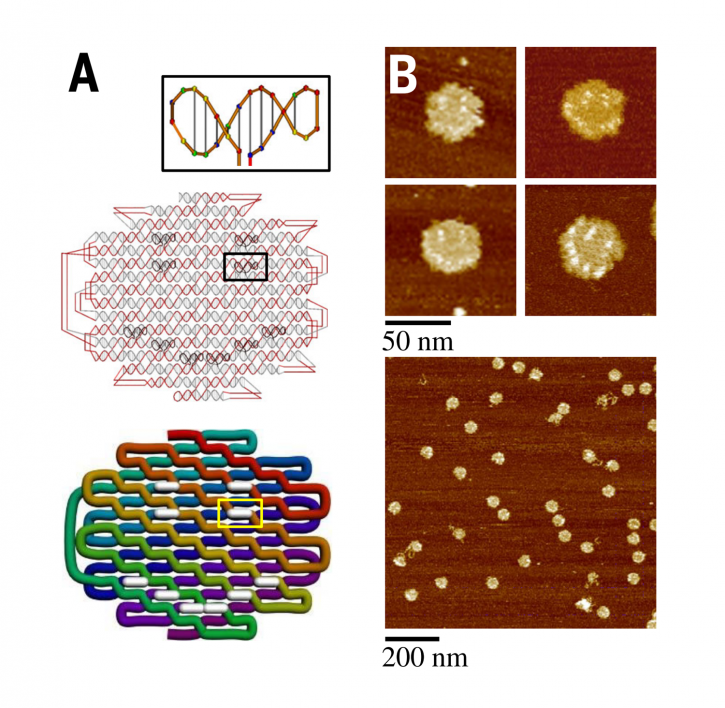
Figure A shows the DNA folding that is designed to self-fold into whatever shapes a scientists can dream up. Figure B shows atomic force microscopy images of emoji-like, nanosized smiley faces.
A new design school
In the software made through a collaboration with BioNano Research Group, Autodesk Research, the user selects a target shape, which is converted into pixelated representation. The user can upload a 2-D image or draw a shape using a 2-D pixel design editor.
The user can optionally add DNA hairpins or loops, which can serve as surface markers or handles for attaching external entities. The pixels are converted into DNA helical domains and locking domains to do the folding. The software will then generate ssOrigami structures and sequences, and the user can view the molecular structure via an embedded molecular viewer. Finally, the DNA sequence is assigned to the cycle strand, and the expected folded structure manufactured in the lab and visually confirmed by viewing it using atomic force microscopy, or AFM.
“We’ve really scaled up the complexity while scaling down the costs,” Yan said. “This study significantly expands the design space and scalability for bottom-up nanotechnology, and opens the door for health applications.”
Top photo: Hao Yan, director of the ASU Biodesign Institute’s Center for Molecular Design and Biomimetics, and the Milton Glick Professor in the School of Molecular Sciences. Photo by Deanna Dent/ASU Now
More Science and technology

ASU-led space telescope is ready to fly
The Star Planet Activity Research CubeSat, or SPARCS, a small space telescope that will monitor the flares and sunspot activity of low-mass stars, has now passed its pre-shipment review by NASA.…

ASU at the heart of the state's revitalized microelectronics industry
A stronger local economy, more reliable technology, and a future where our computers and devices do the impossible: that’s the transformation ASU is driving through its microelectronics research…

Breakthrough copper alloy achieves unprecedented high-temperature performance
A team of researchers from Arizona State University, the U.S. Army Research Laboratory, Lehigh University and Louisiana State University has developed a groundbreaking high-temperature copper alloy…