Top-down design brings new DNA structures to life
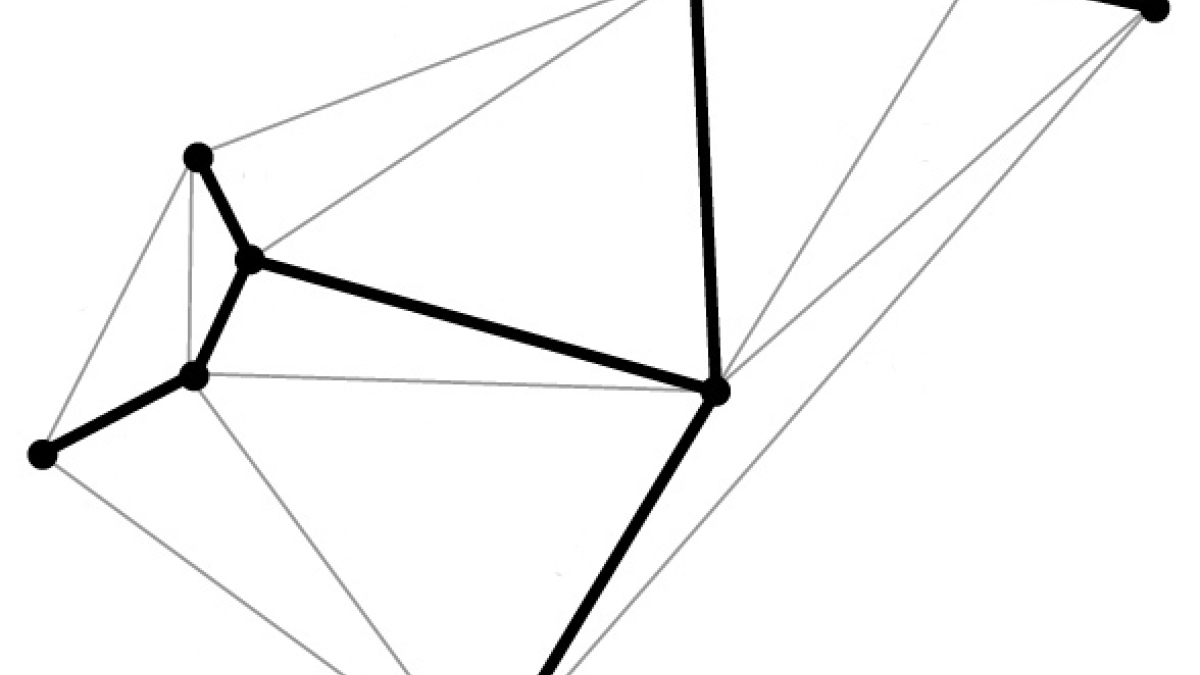
The boldfaced line, known as a spanning tree, follows the desired geometric shape, touching each vertex just once. A spanning-tree algorithm is used in the new DNA origami method to map out the proper routing path for the DNA strand.
Among the valuable holdings in London’s Wellcome Library is a rough pencil sketch made in 1953 by Francis Crick. The drawing is one of the first to show the double-helix structure of DNA — nature’s blueprint for the design of sea snails, human beings and every other living form on Earth.
Few could have predicted however, that DNA’s simple properties of self-assembly and its versatile information-carrying capacity could be put to many uses never imagined by double-helix discoverers James Watson and Crick (or indeed, by nature itself).
In new research appearing in the advance online edition of the journal Science, Arizona State University researcher Hao Yan and colleagues from MIT and Baylor College of Medicine describe a new method for designing geometric forms built from DNA. They present a novel variant on a technique known as DNA origami, in which the base-pairing properties of DNA are exploited for the construction of tiny structures in two and three dimensions.
“An important challenge in the field of DNA nanotechnology is to design any desirable structures in a top-down manner, without much human input concerning details of DNA strand folding paths,” Yan said.
His collaborators at MIT, led by Mark Bathe, developed a computer algorithm to design DNA nanostructures by only inputting a target shape. They engineered a software platform that can compute and output necessary DNA strands to form designer architectures. Formation of these structures were then systematically characterized and confirmed experimentally at the three institutes.
“This really showcases interdisciplinary collaborative science across the country,” said Yan (pictured left), who directs the Biodesign Institute’s Center for Molecular Design and Biomimetics at ASU.
Other worlds
The team designs useful structures at an astonishingly minute scale. (One nanometer is a billionth of a meter, or about the size of a sugar molecule.) Specialized imaging techniques, including atomic force- and cryo-electron microscopy, are used to visualize the resulting forms.
The simplified technique described promises to significantly extend the use of DNA origami beyond the specialist community and expand the range of possible applications in biomolecular science and nanotechnology. These include the use of nanoparticles for drug delivery and cell targeting, construction of nanoscale robots capable of performing diverse activities in medicine and industry and the design of custom-tailored optical devices.
One of the more exciting innovations on the horizon involves the use of DNA as a storage medium — one boasting retention times in the millions of years. (A single gram of DNA can store about 700 terabytes of information — an amount equivalent to 14,000 50-gigabyte Blu-ray disks. Further, such nucleic acid memory could potentially be operated with a fraction of the energy required for other information storage options.)
Shape-shifting
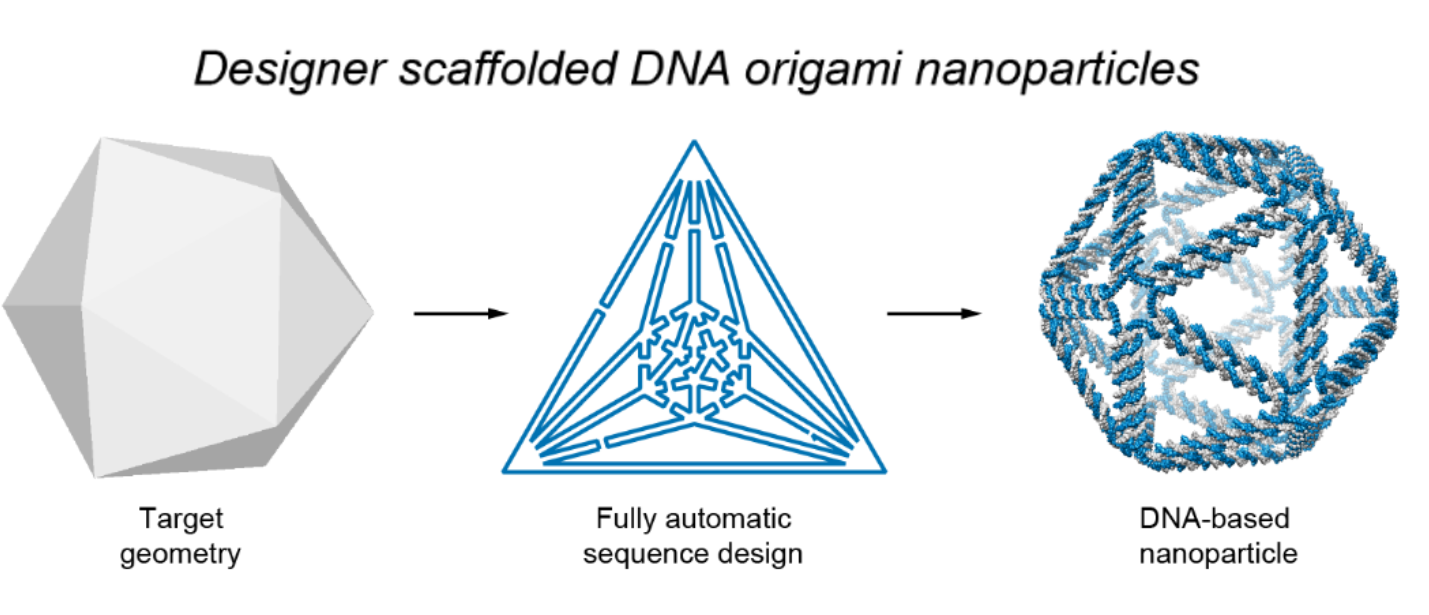
The new design method, which can produce virtually any polyhedral shape, relies on a top-down strategy, which begins with an outline of the desired form and works backward in stages to define the required DNA sequence that will properly fold to form the finished product.
The autonomous process is carried out using a software program designed by the authors. Known as DAEDALUS (for DNA Origami Sequence Design Algorithm for User-Defined Structures), the program carries out inverse design of arbitrary DNA origami nanoforms, based on an input wireframe mesh (a visual representation of the closed, 3-D geometric surface).
Illustration shows the basic process used to design DNA origami nanostructures. First, a wireframe of the intended target design is made. The software then translates this into a plan for the routing of DNA scaffold and staple strands, which assemble to form the desired shape. Image by Biodesign Institute
The program is not only user-friendly, but highly versatile, producing forms not limited to spherical topology (i.e., closed, two-sided structures with no boundaries or holes). Once the target form has been described as a network of nodes and edges, DNA scaffold strands of custom length and sequence are generated using technology known as asymmetric polymerase chain reaction.
The new study describes the fabrication of a variety of geometric DNA objects, including 35 polyhedral forms (Platonic, Archimedean, Johnson and Catalan solids), six asymmetric structures and four polyhedra with nonspherical topology, using inverse design principles. The method can produce nanoforms with high fidelity and stability without the normal laborious process of manually designing base pairs to form the intended target structure.
Entering the fold
DNA origami brings the ancient Japanese method of paper folding down to the molecular scale. The basics are simple: Take a length of single-stranded DNA and guide it into a desired shape, fastening the structure together using shorter so-called staple strands, which bind in strategic places along the longer length of DNA. The method relies on the fact that DNA’s four nucleotide letters — A, T, C and G — stick together in a consistent manner; As always pairing with Ts and Cs with Gs.
The DNA molecule in its characteristic double-stranded form is fairly stiff, compared with single-stranded DNA, which is flexible. For this reason, single-stranded DNA makes for an ideal lace-like scaffold material. Further, its pairing properties are predictable and consistent (unlike RNA, which is considered promiscuous, due to base pairings that may be unexpected).
The technique has proven wildly successful in creating myriad forms in two and three dimensions, which conveniently self-assemble when the designed DNA sequences are mixed together. The tricky part is preparing the proper DNA sequence and routing design for scaffolding and staple strands in order to achieve the desired target structure. Typically, this is painstaking work that must be carried out manually.
With the new technique, the target structure is first described in terms of a wire mesh made up of polyhedra. From this, a spanning tree algorithm is generated. This is basically a map that will automatically guide the routing of the DNA scaffold strand through the entire origami structure, touching each vertex in the geometric form once. Complementary staple strands are then assigned, and the final form self-assembles.
To test the method, simpler forms known as Platonic solids were first fabricated, followed by increasingly complex structures. These included objects with nonspherical topologies and unusual internal details, which had never been experimentally realized before.
The completed designs demonstrated the ability of the top-down technique to automatically generate scaffold and staple routings for an expansive range of nanoforms, based solely on surface geometry. Cryo-EM was used to confirm structural fidelity and stability of the assembled origami structures.
Further experiments confirmed that the DNA structures produced were potentially suitable for biological applications as they displayed long-term stability in serum and low-salt conditions.
The research paves the way for the development of designed nanoscale systems mimicking the properties of viruses, photosynthetic organisms and other sophisticated products of natural evolution.
In addition to his appointment at the Biodesign Institute, Hao Yan is the Milton D. Glick Distinguished Professor in the School of Molecular Sciences, in ASU’s College of Liberal Arts and Sciences.
More Science and technology

ASU-led space telescope is ready to fly
The Star Planet Activity Research CubeSat, or SPARCS, a small space telescope that will monitor the flares and sunspot activity…

ASU at the heart of the state's revitalized microelectronics industry
A stronger local economy, more reliable technology, and a future where our computers and devices do the impossible: that’s the…

Breakthrough copper alloy achieves unprecedented high-temperature performance
A team of researchers from Arizona State University, the U.S. Army Research Laboratory, Lehigh University and Louisiana State…