It turns out that the rigid “line in the sand” between the human sex chromosomes — the Y and X — is a bit blurrier than previously thought.
Contrary to the current scientific consensus, Arizona State University assistant professor Melissa Wilson Sayres has led a research team that has shown that X and Y DNA swapping may occur much more often. And this promiscuous swapping may, in turn, aid in our understanding of human history and diversity, health and disease, as well as blur rigid chromosomal interpretations of sexual identity.
“Studying our sex chromosomes has consequences for human health and for trying to understand our history,” said Melissa Wilson Sayres, an assistant professor in the School of Life SciencesThe School of Life Sciences is an academic unit of the College of Liberal Arts and Sciences. and member of the Biodesign Institute’s Center for Evolution and Medicine. “To me, understanding the evolution of the X and Y is so important because we need to understand that there are all of these variations in the genetics of sex determination.”
Contrary to the current scientific consensus, Arizona State University assistant professor Melissa Wilson Sayres has led a research team that has shown that X and Y DNA swapping may occur much more often.
All of human diversity is spooled within our DNA across 23 pairs of chromosomes, containing an estimated 25,000 genes. For every generation, this DNA information — often including a baby’s sex — is shuffled like a deck of cards between a mother and father through a process called recombination. Recombination makes every individual unique, down to the last pair of sex chromosomes.
Recombination occurs routinely everywhere except on the sex chromosomes, where the genetic deck of cards remains stacked, unable to shuffle information — with the exception of two small regions located at the tips of the X and Y chromosome, called pseudoautosomal regions (PAR1 and PAR2).
“The pseudoautosomal region, this tiny region that still recombines, is extremely understudied, typically filtered out of all analyses,” said Wilson Sayres. In addition, there is a rogue island of the X chromosome, called the X-transposed region, or XTR, which was duplicated from the X to the Y around the last common ancestor of all humans.
The study, published in the early online edition of the journal Genetics, includes ASU School of Life Sciences researchers Daniel J. Cotter and Sarah M. Brotman. Together, they challenged the widely held assumption that genetically there is a strict recombination boundary that suppresses swapping between the X and Y.
Using the complete DNA sequence information from the X chromosomes of 26 unrelated females, the ASU team has shown that the genetic diversity in the region, called PAR1, is far greater than the other regions of the X, and that the diversity is elevated across the PAR1 region, rather than an abrupt cutoff as previously expected. After the PAR1, the diversity should drop off like a cliff, but instead, looks instead like a slow rolling hill, which could result in an increase in the number of X-linked disorders.
To understand the modern X and Y, evolutionary biologists like Wilson Sayres have traced their history back to the dawn of mammals. About 200 million years ago, the X and Y were indistinguishable, but then had a long, drawn-out breakup.
It’s thought that little pieces of the future Y started doing genetic backflips, called inversions, that made it harder to recombine, and the genetic gulf between the sexes first began to widen. In addition to the PAR regions, XTR (X-transposed region) duplicated from the X to the Y in human after the human-chimp split about 6 million years ago, with two genes floating off on this genetic island.
The evolutionary outcome is striking; after 200 million years, the male Y is pruned up, having lost nearly 90 percent of the genes on the ancestral sex chromosomes and the ability to exchange information with the X.
Typically PAR1 and PAR2 are filtered out during sex demographic history studies, while XTR is not. To avoid bias in reporting and interpreting diversity, the new results need to be carefully factored in for future studies.
And understanding the differences between the sex chromosomes is essential for understanding traits involved in some sex-biased diseases (most famously in color-blindness and hemophilia).
For example, a deficiency in PAR1 recombination has been linked to Klinefelter’s syndromeKlinefelter syndrome is a chromosomal condition that affects males, most often resulting from the presence of one extra copy of the X chromosome in each cell. Symptoms can include small testes, decreased facial hair, learning disabilities, delayed speech and a tall, thin body with disproportionately long limbs. (XXY individuals), and what is especially intriguing to Wilson Sayres is what she deems an “unlucky” break involved in a key male reproductive switch, a testis determination region located nearby.
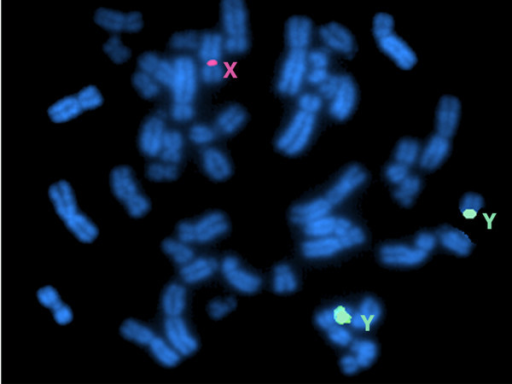
Understanding the differences between the sex chromosomes is essential for understating traits involved in some sex-biased diseases (most famously in color-blindness and hemophilia). The image is a metaphase FISH analysis showing two green signals of the centromere of the Y chromosomes and one red signal of the centromere of the X chromosome confirming the karyotype of XYY. Courtesy of the National Library of Medicine
“This sex-determining region of the Y in the testis determining pathway is now, in humans, right next to the boundary,” said Wilson Sayres. “The big implication is that because of the way our Y chromosome is structured, SRY is immediately next to the boundary, and because the boundary is fuzzy, we can get SRY hopping over to an X chromosome.”
SRY can be shuffled to the X, resulting in an increase in sex-linked disorders, such as SRY-positive XX males, known as de la Chapelle syndromeThe XX male syndrome, also called de la Chapelle syndrome, is a rare disorder in which individuals who have two X chromosomes — a pattern typically found in females — but have a male appearance. People with this disorder have external male genitalia; symptoms include small testes, infertility, short stature and, sometimes, feminine characteristics. According to the National Library of Medicine, in about 80 percent of XX males, the condition results from an abnormal exchange of genetic material in which the SRY gene (usually located on the Y chromosome) is misplaced onto an X chromosome. This form of the condition is SRY-positive XX male syndrome..
“We know that large aspects of gender are really built on societal expectations, but it turns out that also our ideas of what sex is, genetically, may also be a little bit determined by public consensus. Sex has to do with if you are making eggs or sperm in humans. Sex, in fact, can be decoupled from your sex chromosomes. This fuzzy boundary makes it even more messy.”
Other fuzzy sex-linked boundaries include Turner syndromeIn females with this syndrome, all or part of one of the X chromosomes is missing. Symptoms vary and can include a short, stocky build; failure to start puberty; infertility; heart defects; certain learning disabilities and social adjustment problems. (females with only one X), affecting one in 2,500 individuals, and Klinefelter's syndrome, found in one in a 1,000 individuals. Wilson Sayres, who specializes in computational biology, notes that despite what on the surface seem like rare conditions are not so rare if we changed our mind-set.
“Let’s think about the ASU population. We have about 70,000 students, so we expect at least 14 people to have a single X chromosome and 35 to 70 to have two X chromosomes and a Y. Many people may not know their chromosome complement. We should be really careful when we are trying to define someone by their sex chromosomes.”
“There are so many things that can affect sex determination. But (SRY) is the first switch in the testis determining pathway. I call it more like a dimmer. There is another syndrome called Swyer syndrome. An individual will inherit a Y chromosome, from their genetic father, and their SRY gene is turned on, but not on enough to turn on testis determination; they are XY, but develop ovaries.”
Recombination suppression between X and Y is still an actively evolving process in humans. In an intriguing area ripe for future exploration, Wilson Sayres notes that there are 24 additional genes located within PAR1 and countless others near the PAR1 boundary, which have been shown to be important for bone growth, melatonin production, and links to psychiatric disorders, including bipolar affective disorder.
More Science and technology

ASU-led Southwest Advanced Prototyping Hub awarded $21.3M for 2nd year of funding for microelectronics projects
The Southwest Advanced Prototyping (SWAP) Hub, led by Arizona State University, has been awarded $21.3 million in Year 2 funding under the CHIPS and Science Act to continue its work advancing the…

Celebrating '20 Years of Discovery' at the Biodesign Institute
Editor’s note: The Biodesign Institute at Arizona State University wraps up its 20th anniversary with the sixth and final installment of its "20 Years of Discovery" series. Each story highlights…

Student research supports semiconductor sustainability
As microelectronics have become an increasingly essential part of modern society, greenhouse gas emissions, which are associated with their use and manufacture, have increased in tandem.…