Researchers at ASU's School of Molecular Sciences, Biodesign Institute, Mayo Clinic unravel DNA repair mechanism
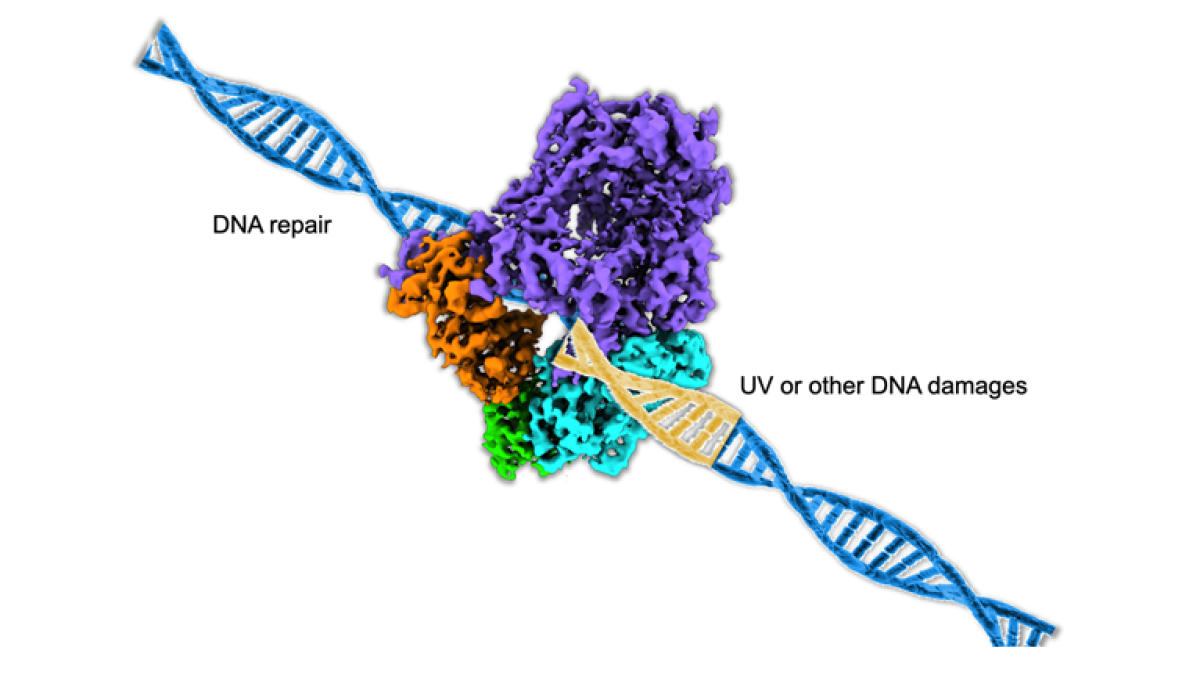
Model of DNA polymerase repairing damaged DNA strand.
The health of a cell is largely determined by the health of its DNA and the genetic information it carries. The correct genetic code will ensure the regular functions to maintain the life of a cell. When DNA becomes damaged, the cell’s repair mechanism, the DNA polymerase enzyme, plays a crucial role in detecting and repairing genetic damage.
DNA can be fragile and damaged by environmental factors, such as chemicals or ultraviolet light. For example, when ultraviolet light damages DNA in skin cells, delicate molecular machinery can detect the damaged gene and repair it. This molecular work is usually accomplished by the collaborative work of proteins. The DNA repair mechanism is robust and requires high accuracy; otherwise, the genetic information that is passed down to the next generation of cells will be incorrect.
In their study, "Cryo-EM reveals conformational flexibility in apo DNA polymerase (zeta)," published in the Journal of Biological Chemistry, Chloe Truong and Po-Lin Chiu of ASU's School of Molecular Sciences and The Biodesign Institute, along with colleagues at Mayo Clinic in Rochester, Minnesota, biochemically characterize the protein-protein interactions of a huge DNA repair enzyme, DNA polymerase zeta, which is responsible for repair across damaged DNA, known as DNA translesion synthesis (TLS).
“This study focuses on solving the structure of polymerase zeta protein complex, which is involved in cell survival from DNA damages,” said Truong, first author of the journal paper.
Using state-of-the-art, high-resolution cryo-EM, or cryogenic electron microscopy, the structure and flexibility of the protein structure were determined. The team recorded millions of electron images of the DNA polymerase zeta and calculated a high-resolution density map by back-projecting these two-dimensional images into a three-dimensional structure of the DNA polymerase zeta. This allowed the team to analyze the molecular flexibility around the section of polymerase zeta responsible for the repair, the DNA binding tunnel. The information gained in this study was used to demonstrate how a cell can fix damaged DNA for continued function and survival.
The molecular images of the DNA polymerase zeta were recorded using the Titan Krios transmission electron microscope hosted in the Eyring Materials Center at ASU.
“Cryo-EM is an excellent tool to learn how protein molecules communicate with each other in a cell,” said Chiu. “This work contributes to understanding how cells survive in vulnerable environments. The structural knowledge of this huge enzyme molecule can provide a framework for developing new cancer therapeutics.”
Tijana Rajh, director of the School of Molecular Sciences, commented, “It is inspiring to see how the cross-pollination of disciplines results in new, important atomic understandings of processes within our cells. This collaborative effort of (the School of Molecular Sciences), The Biodesign Institute and Mayo Clinic was the key to solve another collaborative effort — the collaborative work of proteins involved in DNA repair.”
More Health and medicine
The surprising role of gut infection in Alzheimer’s disease
Arizona State University and Banner Alzheimer’s Institute researchers, along with their collaborators, have discovered a…

ASU, University of Wisconsin partner to empower Black people to quit smoking
Arizona State University faculty at the College of Health Solutions are teaming up with the University of Wisconsin to…

New book highlights physician wellness, burnout solutions
Health care professionals dedicate their lives to helping others, but the personal toll of their work often remains hidden.A new…